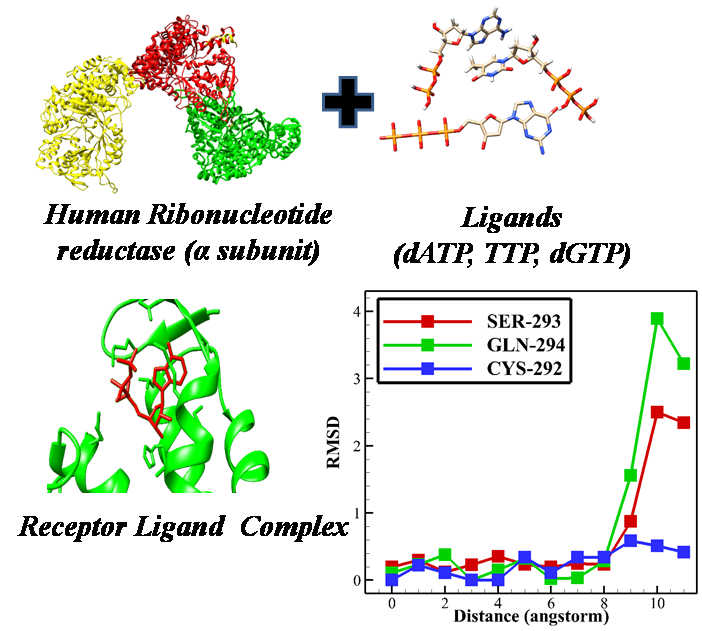
Human Ribonucleotide reductase (RNR) is an essential enzyme that regulates the building blocks of DNA by converting the ribonucleoside diphosphate to deoxyribonucleoside. It is an enzyme essential for maintaining the correct proportions of dNTPs for healthy cellular homeostasis. The disproportionate pool of dNTPs hampers correct replication of cells and leads to mutagenesis, and cell death. In cancerous cells, due to high cell breeding, dNTPs pools are low, which leads to replication stress and further genomic instability. Responsible residues that maintain this balance of dNTPs have not been recognized yet. We explored how three different ligands (ATP, dTTP, and dGTP) to the substrate specificity site of the enzyme and the residues that hold the ligand in place and found that similar to other class 1 RNRs a residue filters which ribonucleotide diphosphate based on its size. Results showed that dGTP is much more strongly attached to the substrate specificity site than TTP. These results give a qualitative understanding of the allosteric mechanism of human RNR and can be helpful in designing smart medicines for cancer treatment.